Alexander Joo-Hyun Sullivan
Software Developer & Bioinformatician
Featured Projects
Featured Publications
20 years of the Bio-Analytic Resource for Plant Biology
Alexander Sullivan, Michael N Lombardo, Asher Pasha, Vincent Lau, Jian Yun Zhuang, Ashley Christendat, Bruno Pereira, Tianhui Zhao, Youyang Li, Rachel Wong, Faisal Z Qureshi, Nicholas J Provart
10.1093/nar/gkae920 | Nucleic Acids Research | 2024-10-23
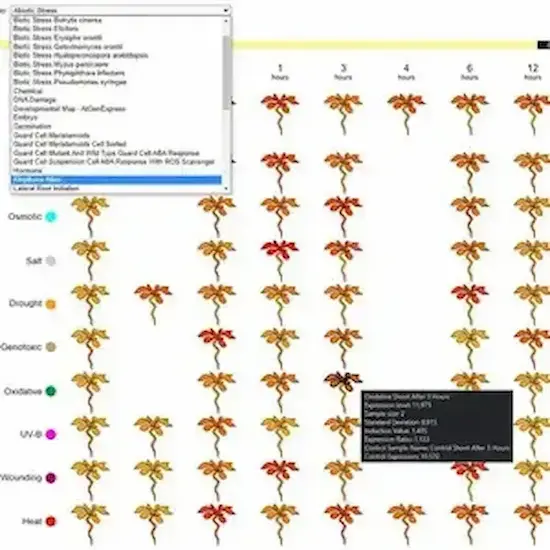
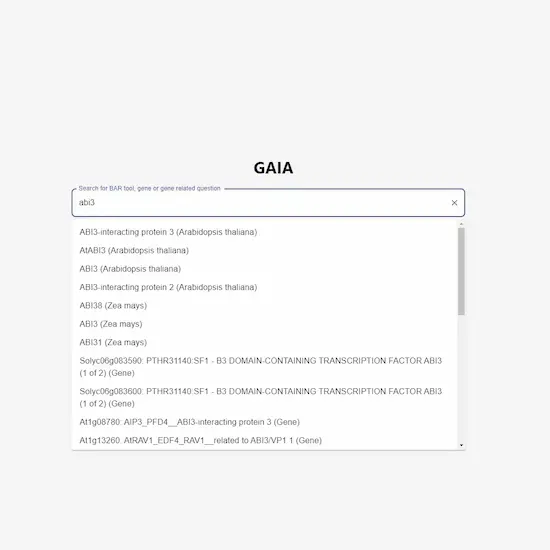
The Bio-Analytic Resource for Plant Biology (‘the BAR’, at https://bar.utoronto.ca) is celebrating its 20th year in operation in 2025. The BAR encompasses and provides visualization tools for large ‘omics data sets from plants. The BAR covers data from Arabidopsis, tomato, wheat, barley and 29 other plant species (with data for 2 others to be released soon). These data include nucleotide and protein sequence data, gene expression data, protein-protein and protein–DNA interactions, protein structures, subcellular localizations, and polymorphisms...
ePlant in 2021: New Species, Viewers, Data Sets, and Widgets
Ben Waese-Perlman, Asher Pasha, Chantal Ho, Amirahmad Azhieh, Yushan Liu, Alexander Sullivan, Vincent Lau, Eddi Esteban, Jamie Waese, George Ly, Cornelia Hooper, S. Evan Staton, Nicholas Brereton, Cuong Le, Rex Nelson, Shelley Lumba, David Goodstein, A. Harvey Millar, Isobel Parkin, Lewis Lukens, Juergen Ehlting, Loren Rieseberg, Frédéric Pitre, Anne Brown, Nicholas J. Provart
10.1101/2021.04.28.441805 | Preprint (bioRxiv) | 2021-04-29
ePlant was introduced in 2017 for exploring large Arabidopsis thaliana data sets from the kilometre to nanometre scales. In the past four years we have used the ePlant framework to develop ePlants for 15 agronomically-important species: maize, poplar, tomato, Camelina sativa, soybean, potato, barley, Medicago truncatula, eucalyptus, rice, willow, sunflower, Cannabis sativa, wheat and sugarcane. We also updated the interface to improve performance and accessibility, and added two new views to the Arabidopsis ePlant - the Navigator and Pathways v...
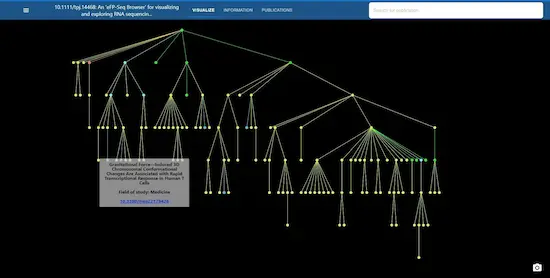
An 'eFP-Seq Browser' for visualizing and exploring RNA sequencing data
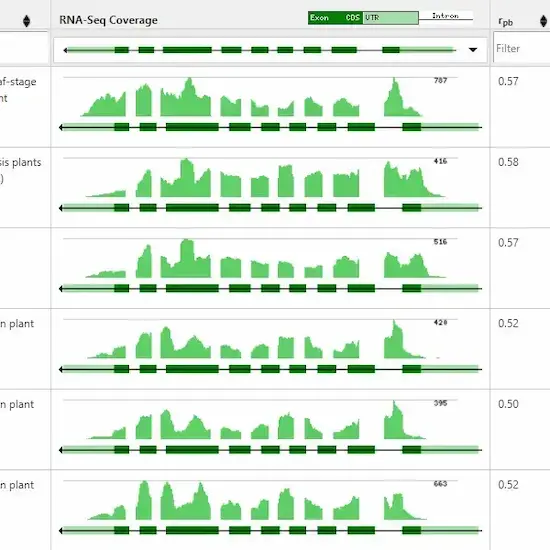
Alexander Sullivan, Priyank Purohit, Nowlan H. Freese, Asher Pasha, Eddi Esteban, Jamie Waese, Alison Wu, Michelle Chen, Chih Ying Chin, Richard Song, Snehal R. Watharkar, Agnes P. Chan, Vivek Krishnakumar, Matthew W. Vaughn, Chris Town, Ann E. Loraine, Nicholas J. Provart
10.1111/tpj.14468 | The Plant Journal | 2019-07-26
Improvements in next-generation sequencing technologies have resulted in dramatically reduced sequencing costs. This has led to an explosion of '-seq'-based methods, of which RNA sequencing (RNA-seq) for generating transcriptomic data is the most popular. By analysing global patterns of gene expression in organs/tissues/cells of interest or in response to chemical or environmental perturbations, researchers can better understand an organism's biology. Tools designed to work with large RNA-seq data sets enable analyses and visualizations to help...